|
Group Publications
*indicates corresponding author |
2024
|
Chung KP, Frieboese D, Waltz F, Engel BD, Bock R* (2024). Identification and characterization of the COPII vesicle-forming GTPase Sar1 in Chlamydomonas. Plant Direct. 8:e614.
|
|
Yamauchi KA , Lamm L, Gaifas L, Righetto RD, Litvinov D, Engel BD*, Harrington K* (2024). Surforama: interactive exploration of volumetric data by leveraging 3D surfaces. bioRxiv. 2024.05.30.596601.
|
|
McCafferty CL*, Klumpe S*, Amaro RE*, Kukulski W*, Collinson L*, Engel BD* (2024). Integrating cellular electron microscopy with multimodal data to explore biology across space and time. Cell. 187: 563-584.
|
|
Lamm L, Zufferey S, Righetto RD, Wietrzynski W, Yamauchi KA, Burt A, Liu Y, Zhang H, Martinez-Sanchez A, Ziegler S, Isensee F, Schnabel JA, Engel BD*, Peng T* (2024). MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography. bioRxiv. 2024.01.05.574336.
|
2023
|
Shimakawa G , Demulder M, Flori S, Kawamoto A, Tsuji Y, Nawaly H, Tanaka A, Tohda R, Ota T, Matsui H, Morishima N, Okubo R, Wietrzynski W, Lamm L, Righetto RD, Uwizeye C, Gallet B, Jouneau P-H, Gerle C, Kurisu G, Finazzi G, Engel BD*, Matsuda Y* (2023). Diatom pyrenoids are encased in a protein shell that enables efficient CO2 fixation. bioRxiv. 2023.10.25.564039.
|
|
Bregy I, Radecke J, Noga A, van den Hoek H, Kern M, Haenni B, Engel BD, Siebert CA, Ishikawa T, Zuber B*, Ochsenreiter T* (2023). Cryo-electron tomography sheds light on the elastic nature of the Trypanosoma brucei tripartite attachment complex. bioRxiv. 2023.03.06.531305.
|
|
Wietrzynski W*, Engel BD* (2023). Supramolecular organization of chloroplast membranes. The Chlamydomonas Sourcebook (Third Edition). Volume 2: Organellar and Metabolic Processes. 2: 763-785.
|
|
Goodenough U*, Engel BD* (2023). Cell Ultrastructure. The Chlamydomonas Sourcebook (Third Edition). Volume 1: Introduction to Chlamydomonas and its Laboratory Use. 1: 17-40.
|
2022
|
van den Hoek H, Klena N, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan W, Geimer S, Plitzko JM, Baumeister W, Pigino G*, Hamel V*, Guichard P*, Engel BD* (2022). In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Science. 377: 543-548.
|
|
Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Trischler R, Schuller SK, Wagner J, Schwarz FM, Engel BD*, Müller V*, Schuller JM* (2022). Membrane-anchored HDCR nanowires drive hydrogen-powered CO2 fixation. Nature. 607: 823-830.
|
|
Lamm L, Righetto RD, Wietrzynski W, Pöge M, Martinez-Sanchez A, Peng T*, Engel BD* (2022). MemBrain: a deep learning-aided pipeline for automated detection of membrane proteins in cryo-electron tomograms. Computer Methods and Programs in Biomedicine. 224: 106990.
|
|
Righetto RD*, Engel BD* (2022). Expanding the arsenal of bacterial spearguns. Nature Microbiology. 7: 363–364.
|
2021
|
Waltz F, Salinas-Giegé T, Englmeier R, Meichel H, Soufari H, Kuhn L, Pfeffer S, Förster F, Engel BD, Giegé P*, Drouard L*, Hashem Y* (2021). How to build a ribosome from RNA fragments in Chlamydomonas mitochondria. Nature Communications. 12: 7176.
|
|
Moebel E, Martinez-Sanchez A, Lamm L, Righetto RD, Wietrzynski W, Albert S, Larivière D, Fourmentin E, Pfeffer S, Ortiz J, Baumeister W, Peng T, Engel BD*, Kervrann C* (2021). Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nature Methods. 18: 1386–1394.
|

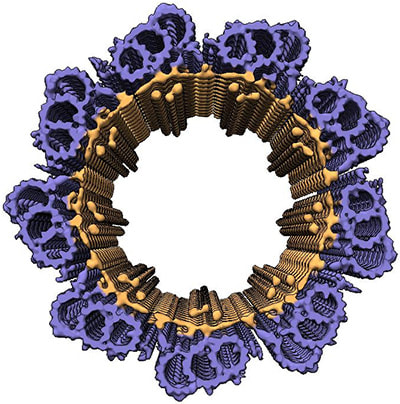
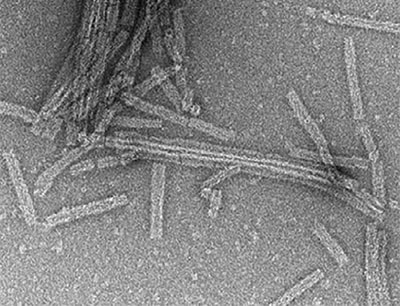
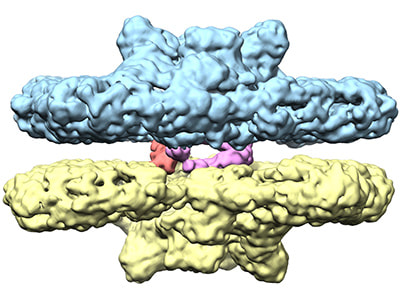
Gupta TK, Klumpe S, Gries K, Heinz S, Wietrzynski W, Ohnishi N, Niemeyer J, Spaniol B, Schaffer M, Rast A, Ostermeier M, Strauss M, Plitzko JM, Baumeister W, Rudack T, Sakamoto W, Nickelsen J, Schuller JM*, Schroda M*, Engel BD* (2021). Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity. Cell. 184: 3643-3659.e23.
|
|
Wietrzynski W*, Engel BD* (2021). Chlorophyll biogenesis sees the light. Nature Plants. 7: 380–381.
|
|
Zabret J, Bohn S, Schuller SK, Arnolds O, Möller M, Meier-Credo J, Liauw P, Chan A, Tajkhorshid E, Langer JD, Stoll R, Krieger-Liszkay A, Engel BD, Rudack T*, Schuller JM*, Nowaczyk MM* (2021). Structural insights into photosystem II assembly. Nature Plants. 7: 524-538.
|
2020
|
He S, Chou H-T, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, He G, Chen VK, Hughson FM, McCormick AJ, Mueller-Cajar O, Engel BD, Yu Z, Jonikas MC* (2020). The structural basis of Rubisco phase separation in the pyrenoid. Nature Plants. 6: 1480–1490.
|

Klena N, Le Guennec M, Tassin A-M, van den Hoek H, Erdmann PS, Schaffer M, Geimer S, Aeschlimann G, Kovacik L, Sadian Y, Goldie KN, Stahlberg H, Engel BD*, Hamel V*, Guichard P* (2020). Architecture of the centriole cartwheel-containing region revealed by cryo-electron tomography. EMBO Journal. 39: e106246.
|
|
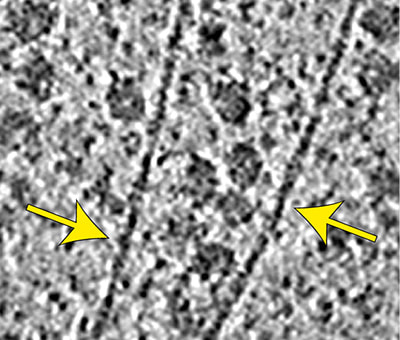
Wietrzynski W, Schaffer M, Tegunov D, Albert S, Kanazawa A, Plitzko JM, Baumeister W, Engel BD* (2020). Charting the native architecture of Chlamydomonas thylakoid membranes with single-molecule precision. eLife. 9: e53740.
|
|
Le Guennec M, Klena N, Gambarotto D, Laporte MH, Tassin AM, van den Hoek H, Erdmann PS, Schaffer M, Kovacik L, Borgers S, Goldie KN, Stahlberg H, Bornens M, Azimzadeh J, Engel BD*, Hamel V*,Guichard P* (2020). A helical inner scaffold provides a structural basis for centriole cohesion. Science Advances. 6: eaaz4137.
|
|
Theis J, Niemeyer J, Schmollinger S, Ries F, Rütgers M, Kumar Gupta T, Sommer F, Muranaka LS, Venn B, Schulz-Raffelt M, Willmund F, Engel BD, Schroda M* (2020). VIPP2 interacts with VIPP1 and HSP22E/F at chloroplast membranes and modulates a retrograde signal for HSP22E/F gene expression. Plant, Cell & Environment. 43: 1212-1229.
|

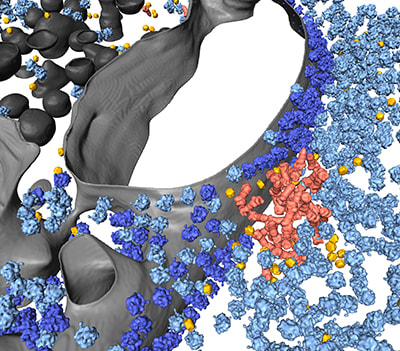
Albert S, Wietrzynski W, Lee CW, Schaffer M, Beck F, Schuller JM, Salomé PA, Plitzko JM, Baumeister W*, Engel BD* (2020). Direct visualization of degradation microcompartments at the ER membrane. Proceedings of the National Academy of Sciences USA. 117: 1069-1080.
|
2019
|
Craig EW, Mueller DM, Bigge BM, Schaffer M, Engel BD, Avasthi P* (2019). The elusive actin cytoskeleton of a green alga expressing both conventional and divergent actins. Molecular Biology of the Cell. 30: 2827-2837.
|
|
Schaffer M*, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, Plitzko JM* (2019). A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nature Methods. 16: 757-762.
|
|
Theis J, Kumar Gupta T, Klingler J, Wan W, Albert S, Keller S*, Engel BD*, Schroda M* (2019). VIPP1 rods engulf membranes containing phosphatidylinositol phosphates. Scientific Reports. 9: 8725.
|
|
Rast A, Schaffer M, Albert S, Wan W, Pfeffer S, Beck F, Plitzko JM, Nickelsen J*, Engel BD* (2019). Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nature Plants. 5: 436-446.
|
|
Schuller JM*, Birrell JA, Tanaka H, Konuma T, Wulfhorst H, Cox N, Schuller SK, Thiemann J, Lubitz W, Sétif P, Ikegami T, Engel BD, Kurisu G*, Nowaczyk MM* (2019). Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science. 363: 257-260.
|
2018
|
Kovtun O, Leneva N, Bykov YS, Ariotti N, Teasdale RD, Schaffer M, Engel BD, Owen DJ, Briggs JAB*, Collins BM* (2018). Structure of the membrane-assembled retromer coat by cryo-electron tomography. Nature. 561: 561-564.
|

Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, Chung JK, Plitzko JM, Groves JT, Jacobs-Wagner C, Engel BD*, Holt LJ* (2018). mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell. 174: 338-349.e320.
|
|
Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD*, Beck M* (2018). In situ architecture of the algal nuclear pore complex. Nature Communications. 9: 2361.
|
2017
|
Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W*, Engel BD* (2017). Proteasomes tether to two distinct sites at the nuclear pore complex. Proceedings of the National Academy of Sciences USA. 114: 13726-13731.
|
Bykov YS, Schaffer M, Dodonova SO, Albert S, Plitzko JM, Baumeister W*, Engel BD*, Briggs JAG* (2017). The structure of the COPI coat determined within the cell. eLife. 6: e32493.
|
Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Förster F, Wingreen NS*, Engel BD*, Mackinder LCM, Jonikas MC* (2017). The Eukaryotic CO2-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Cell. 171: 148-162.e19.
|
|
Albanese P, Melero R, Engel BD, Grinzato A, Berto P, Manfredi M, Chiodoni A, Vargas J, Sorzano CÓS, Marengo E, Saracco G, Zanotti G, Carazo JM, Pagliano C* (2017). Pea PSII-LHCII supercomplexes form pairs by making connections across the stromal gap. Scientific Reports. 7: 10067.
|
|
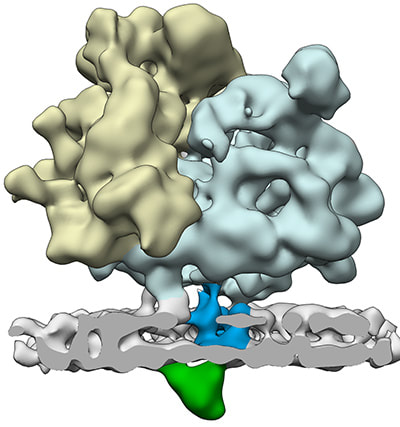
Pfeffer S, Dudek J, Schaffer M, Ng BG, Albert S, Plitzko JM, Baumeister W, Zimmermann R, Freeze HH, Engel BD*, Förster F* (2017). Dissecting the molecular organization of the translocon-associated protein complex. Nature Communications. 8: 14516.
|
|
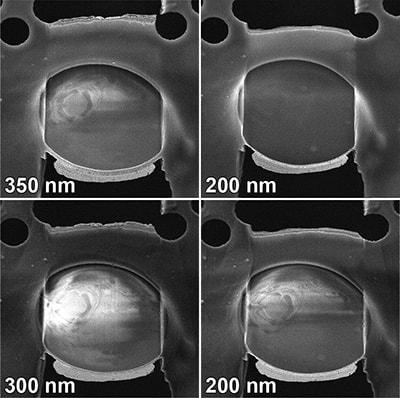
Schaffer M*, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko J* (2017). Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. Journal of Structural Biology. 197: 73-82.
|
2016
|
Asano S, Engel BD, Baumeister W* (2016). In situ cryo-electron tomography: a post-reductionist approach to structural biology. Journal of Molecular Biology. 428: 332-343.
|
2015
|
Engel BD*, Schaffer M, Albert S, Asano S, Plitzko JM, Baumeister W* (2015). In situ structural analysis of Golgi intracisternal protein arrays. Proceedings of the National Academy of Sciences USA. 112: 11264-11269.
|
|
Schaffer M*, Engel BD, Laugks T, Mahamid J, Plitzko JM, Baumeister W (2015). Cryo-focused ion beam sample preparation for imaging vitreous cells by cryo-electron tomography. Bio-protocol. 5: e1575.
|
|
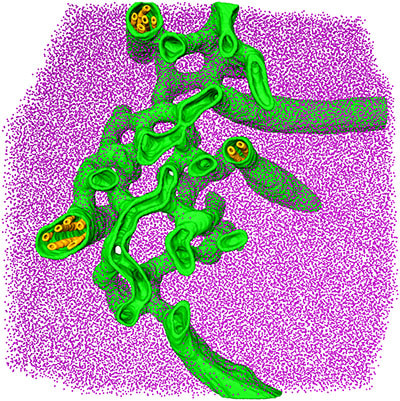
Engel BD*, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W* (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife. 4: e04889.
|